Ubiquitination
1.1 Ubiquitin
First identified in 1975, ubiquitin is a regulatory protein that is highly conserved throughout eukaryotes. It is a small protein consisting of 76 amino acids with a molecular weight of 8.5 kDa. Its main features are its C-terminal tail and lysine residues. Ubiquitin is found in both the cytoplasm where it is involved in ATP-dependent proteolysis pathway and the nucleus where it may play a role in regulation of chromatin structure and/or regulation of transcriptional activity (Lund, 1985; Goldknopf, 1977; Levinger, 1982).
1.2 Ubiquitination
Ubiqitination is a process first identified in the 1980s that gained Nobel Prize recognition for Aaron Ciechanover, Avram Hershko and Irwin Rose in 2004 (Ciechanover, 1980). It is an important and widespread post-translational modification of proteins that resembles phosphorylation. Ubiquitination involves the covalent attachment of ubiquitin to a substrate protein via lysine residues. Ubiquitin itself contains seven conserved lysine residues each capable of isopeptide linkage. This can be in the form of monoubiquitination or polyubiquitination. Ubiquitin signals, or degrons, are usually determined by short regions in the primary sequence of the targeted protein (Bachmair, 1989).
In monoubiquitination, a single ubiquitin moiety is attached to one or several individual lysine residues in the substrate, regulating location and activity of many cellular proteins. In polyubiquitination, ubiquitin molecules are thought to be sequentially added first to a lysine residue of the target, and then to the distal end of the growing ubiquitin chain (Chau, 1989). Lysines K6, K11, K27, K29, K33, K48 and K63 of ubiquitin can potentially form ubiquitin-ubiquitin linkages (Hicke, 2003). The most characterised polyubiquitin chain is the linkage of ubiquitin carboxy-terminal glycines to Lys48 of the preceding ubiquitin which ultimately results in proteasomal degradation (Thrower, 2000). Similarly, Lys29 has also been described as having a role in proteasomal degradation (Rao, 2002). Lys63-linkage has been associated with activation of specific proteins for DNA repair, signal transduction and endocytosis without proteasomal involvement (Pickart, 2004; Sun, 2004). Moreover, these two polyubiquitin chains have been shown to have different conformations, the Lys63 chain being more extended than Lys48 through a different form of ubiquitin linkage (423 Pickart, 2004). Figure 1.1 shows examples of ubiquitin conjugation.
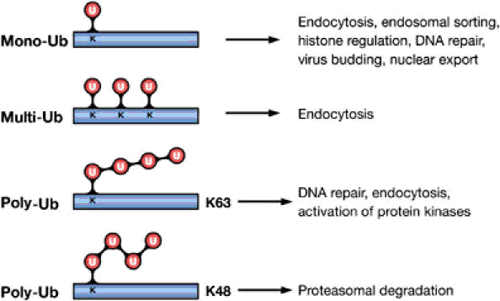
Figure 1.1 Ub modifications and their cellular functions. Attachment of a single Ub molecule to a single Lys (K) residue leads to protein monoubiquitylation (mono-Ub). Multiple monoubiquitylation (multi-Ub) results from the addition of several single Ub molecules to different Lys residues. These modifications are implicated in various nonproteolytic cellular functions, including endocytosis, endosomal sorting and DNA repair. Polyubiquitylation (poly-Ub) results from the attachment of a chain of Ub molecules to one or more Lys residues. Ub chains formed via Lys48 (K48) of Ub target-modified proteins for proteasomal degradation. Chains linked via Lys63 (K63) are implicated in DNA repair and activation of protein kinases (Haglund, 2005).
1.3 The Role of Ubiquitination
Ubiquitination essentially has two functional categories, proteolytic and nonproteolytic. The proteolytic ubiquitination involves tagging target proteins with ubiquitin moieties in a specific manner that is then recognized by the proteasome where the target undergoes proteasomal degradation. Alternatively, targets can be ubiquitinated in an inducible and reversible manner with a role cell signalling; gene transcription, DNA repair and replication, intracellular trafficking and virus budding and replication (reviewed Haglund, 2005; Hochstrasser, 2006).
1.4 The Ubiquitin Pathway
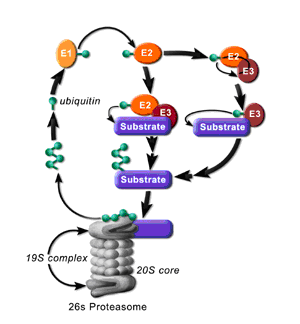
Ubiquitination involves the marking of a target protein with one or more ubiquitin moieties. The ubiquitin marking process occurs in a series of steps known as the ubiquitin pathway that requires three main enzymes. The first is the ubiquitin activation enzyme, E1, which carries out the ATP-dependent activation of the C-terminus of ubiquitin. The ubiquitin molecule then becomes covalently linked through a thioester bond to the active site cysteine of E1. Ubiquitin is next transferred to the active site cysteine of the ubiquitin-conjugating enzyme, E2. Finally, the ubiquitin-protein ligase, E3, catalyses the transfer of ubiquitin to, and thioester bond formation with a lysine residue in the protein substrate or of another ubiquitin already linked to the substrate (Hershko, 1998; Amerik, 2004). Depending on the type of ubiquitination the substrate may be degraded by the proteasome (Figure 1.2) or become part of a cellular signalling network.
 1.4.1 Ubiquitin-proteolytic Pathway: Proteasomes are found in the nucleus and cytoplasm. Their ability to degrade cellular proteins is utilised for abnormal or misfolded proteins in the cell’s stress response process, in cellular differentiation, generating antigenic peptides for the immune system and in regulation of the cell cycle. Most of these processes are related to the ubiquitin pathway. Removal of proteasome genes in the cells is lethal, hence they are essential for cell survival (Ciechanover, 1994).
1.4.1 Ubiquitin-proteolytic Pathway: Proteasomes are found in the nucleus and cytoplasm. Their ability to degrade cellular proteins is utilised for abnormal or misfolded proteins in the cell’s stress response process, in cellular differentiation, generating antigenic peptides for the immune system and in regulation of the cell cycle. Most of these processes are related to the ubiquitin pathway. Removal of proteasome genes in the cells is lethal, hence they are essential for cell survival (Ciechanover, 1994).
Following the ubiquitination process different fates await the substrates. The most characterised polyubiquitin chain which is recognized by the proteasome is the Lys48-linked ubiquitin chain that must consist of at least 4 ubiquitin moieties (Ciechanover, 1994). The proteasome holoenzyme, also known as the 26S proteasome, is a 2.5 mDa complex. It is made up of two copies each of at least 32 different subunits with trypsin, chymotrypsin and peptidylglutamyl peptidase-like activities that are highly conserved among all eukaryotes. The overall structure can be divided into two major sub-complexes. The first is the 20S core particle (CP) that contains the protease subunits where the proteins are actually cleaved and the second is the 19S regulatory particle (RP) that facilitates entry of the proteins into, and regulates the function of, the 20S CP (Rivett, 1993; Tanaka, 1997; Glickman, 2001). The ubiquitin signal is thought to be recognized by receptors such as Rad23 and Dsk2, which contain a ubiquitin-like (UBL) domain that is recognized by the proteasome, and ubiquitin-associated (UBA) domains which bind ubiquitin (Elsasser, 2004; Wilkinson, 2001; Rao, 2002). Upon recognition of the ubiquitin signal the substrate is incorporated into the proteasome where it is hydrolyzed to small peptides and amino acids that leave the catalytic chamber through small pores in the walls of the 20S particle.
Ubiquitination is now also emerging as a key regulator of eukaryotic transcription. One method is as a result of proteasomal degradation of transcription factors. A second method is through mechanisms that involve the ubiquitination and proteolytic processing of proteins from inactive to active transcription factors. An example of this is the nuclear factor kappa B1, NF-κB1. The COOH-terminal domains of the p105 and p100 precursors are ubiquitinated possibly by the SCFβ-TRCP E3 ligase and cleaved by the proteasome to produce the active NF-κB1 p52 and p50 subunits. The subunits are then free to unite with their Rel partners and reconstitute the active NF-κB heterodimer transcription factor (Orian, 1999; Lin, 1996; Heissmeyer, 2001).
The transcription activation domains (TAD) of transcription factors and intact GAL4 DNA binding domains have been shown to be important in the ubiquitination process. Estrogen receptor ligand-dependent transactivation was shown to fail to occur in the absence of its E1 ligase or when the proteasome was chemically inhibited (Lonard, 2000) and Salghetti et al. demonstrated that ubiquitination was required for both the quick turnover of the LexA-VP16 chimeric transcription factor and its transcriptional activity (Salghetti, 2000). This suggests the possibility that ubiquitination plays a dual role in activating transcription factors and then signalling their destruction, although this has not been proven (Figure 1.3) (Conaway, 2002).
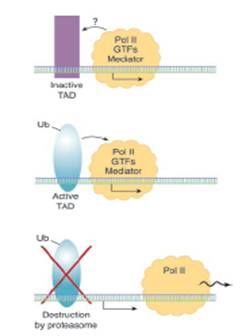 Figure 1.3 – Model of ubiquitination role in transcription. The activation and subsequent destruction of TAD transcription factors. Binding of some transcription factors to promoters triggers their ubiquitination which simultaneously activates them to initiate transcription and tags them for subsequent degradation. {{413 Conaway, 2002; }}
Figure 1.3 – Model of ubiquitination role in transcription. The activation and subsequent destruction of TAD transcription factors. Binding of some transcription factors to promoters triggers their ubiquitination which simultaneously activates them to initiate transcription and tags them for subsequent degradation. {{413 Conaway, 2002; }}
1.4.2 Non-proteolytic Ubiquitination
Monoubiquitination may have several regulatory roles including alteration of the structure, function and/or cellular localisation of the sustrate (Sigismund, 2004; Haglund, 2003; Hicke, 2003). It is now established that monoubiquitination of histones results in heterochromatin relaxation and assembly of transcription complexes on the promoter in a proteasome-independent manner (Spencer, 1999; Pham, 2000; Robzyk, 2000). Monoubiquitination of certain receptor proteins on the plasma membrane can also promote their internalization and down-regulation in a proteasome-independent manner. Ubiquitin signals their endocytic transport and the proteins are targeted for degradation by the lysosome (Hicke, 1999; Strous, 1996; Bonifacino, 1998).
1.4.2.1 Deubiquitination
Genome sequencing projects have identified more than 90 deubiquitinating enzymes (DUB). DUBs have been described as negative regulators of the ubiquitin/proteasome pathway. Believed to reside in the 19S regulatory particle of the 26S proteasome, there are five sub-families of DUBs. These are categorised according to their conserved domains and include the Ubiquitin-Specific Processing Proteases (USP), Ubiquitin Carboxy-terminal Hydrolases (UCH), Ovarian Tumour-related Proteases (OTU), Josephin Domain Proteases and the JAMM/MPN+ Proteases (Turcu, 2010). DUBs are thought to be highly specific to their substrate with different DUBs acting against different lysine residues to cleave ubiquitin. This includes various different types of lysine bonds between ubiquitins distal to the substrate, and bonds between the substrate and ubiquitin. The bonds may be for example cleaved before ubiquitin brings the substrate to the proteasome in order to prevent proteolysis or once the ubiquitin/substrate has met with the proteasome so that the substrate can be degraded without ubiquitin being destroyed (Amerik, 2004; Soboleva, 2004). Recent evidence has suggested, however, that the ubiquitin chain is degraded with the target protein. Ubiquitin has a long half-life but is only required to guide substrates for a short period therefore the ability of DUBs to cleave ubiquitin into monomers to replenish ubiquitin pools and hence be recycled may mean they are the main regulators of ubiquitin in the cell (D’Andrea, 1998; Kim, 2003). The USP family of DUBs have been shown to interact with a range of proteins including the Von Hippel-Lindau tumour suppressor protein pVHL, the hypoxia inducible transcription factor HIF-1α, and the breast cancer 2 gene product BRCA2 (reviewed de Jong, 2006).
USP7 has been shown to stabilize p53, consequently affecting cell viability and apoptosis. The mode of action has not been unequivocally established as USP7 was initially thought to directly interact with p53 and subsequently deubiquitinate it, but studies have also shown that USP7 deubiquitinates MDM2, the E3 ligase for p53 (Li, 2002; Hu, 2002). DUB-3, a cytokine-regulated deubiquitinating enzyme, has been shown to be induced in response to interleukin-4 and interleukin-6 stimulation (Burrows, 2004). These examples demonstrate the importance of deubiquitinating enzymes in the cell cycle.