An investigation into the potential of Geobacter sulfurreducens to encode transglutaminase and its potential for industry.
Abstract
Transglutaminase is a cross-linking enzyme that links proteins to form a covalent bond resistant to proteolytic degradation. In 2009, Janet B. Rollefson found a gene encoding for a putative transglutaminase in Geobacter sulfurreducens, where it was found to be critical for biofilm formation on electrodes in microbial fuel cells. If this enzyme is a transglutaminase, it will be a novel transglutaminase that could be patented, commercialised and marketed. It could be used to replace existing transglutaminases in various applications in the food, medicine, cosmetic and textile industries. It could potentially have other beneficial properties, such as improved activity, altered substrate specificity and/or stability. Investigation of the nature of this enzyme will also be important for optimisation of the growth of G. sulfurreducens in microbial fuel cells. We carried out this study to investigate the potential of this organism as an alternative source of transglutaminase.
Using suitable primers, we amplified this putative transglutaminase gene encoding for both mature and signal peptides containing protein. Thermal cycling conditions were optimised for maximum amplification yield and specificity. We subcloned this into pET21d vector since it has unique restriction sites of Nhe1 and Xho1 and also carries a His tag sequence that simplifies the purification of our ‘TG’ protein. The digested products were ligated together and transformed into BL21(D3) competent E. coli cells. Transformed cells were inducted with 1mM IPTG for 3 hours at 37°C. Analysis of SDS-PAGE revealed the induced protein of the expected size (52 kDa) in Clone3. This clone was harvested, lysed by sonication and cell debris removed by centrifugation. The lysate was applied to a 1 ml His-Bind column. The putative transglutaminase protein was eluted in a linear gradient using imidazole, at a flow rate of 1 ml/min and fractions were collected into a 96 well plate. SDS-PAGE gel electrophoresis of the fraction collected during IMAC purification showed expected bands affirming the presence of a 52kDa protein. Samples were then purified using PD-10 Desalting columns (GE Healthcare) Gravity protocol. Protein samples were collected, marking the end of the expression and purification of the recombinant protein.
Characterisation of this putative transglutaminase was carried out using biochemical assays for transglutaminase. In order to quantify our protein, we carried out the Lowry protein assay using Bio-Rad Dc Protein Assay Reagent A and Reagent B in a 96 well plate. A standard curve of absorbance at 750nm against protein concentration was plotted and the protein concentration in our purified protein samples was found to be 1mg/ml. Transglutaminase assay carried out using 50mM Tris-HCl, 0.25mM biotin cadaverine, 10mM DTT, 20mM CaCl2 and absorbance was read at 490nm. Using mTG as a positive control it was observed that our recombinant protein does not show transglutaminase activity.
Microbial transglutaminase has broad substrate reactivity and is known to cross-link casein to form high molecular weight products. No casein cross-linking was observed using our sample‘TG’ with increasing time whereas microbial transglutaminase cross-links casein, thus showing negative transglutaminase activity.
Thus, Geobacter sulfurreducens does not encode a transglutaminase.
Introduction
Transglutaminase is an enzyme that has commercial importance. Interest in this enzyme is increasing as it has various applications. It is a cross-linking enzyme that links proteins to form a covalent bond resistant to proteolytic degradation. A gene encoding for a putative transglutaminase has been found in Geobacter sulfurreducens(Janet B. Rollefson, 2009). In this research project, we investigate this putative transglutaminase from Geobacter sulfurreducens. If this enzyme is a transglutaminase (as proposed) it will be a novel transglutaminase that could be patented, commercialised and marketed. It could be used to replace mammalian transglutaminase in various existing applications in the food, medicine, cosmetic and textile industries. It could potentially have other beneficial properties, such as improved activity, altered substrate specificity and/or stability. An enzyme from this source could be more thermostable and also increase the rate of reaction. Investigation of the nature of this enzyme will also be important for optimisation of the growth of G. sulfurreducens in microbial fuel cells. The potential of this organism as an alternative source of transglutaminase motivated this study.
Transglutaminase
History and major milestones:
The major milestones in the history if transglutaminase are the following:
- In 1948, Factor XIII (fibrin stabilising factor) was discovered by Laki-Lorand.
- In 1957, transglutaminase activity was observed in guinea pig liver (Clarke D. D, 1957).
- It was observed that transglutaminase also played in a role in blood clotting by stabilising the fibrin monomers (Pisano, 1968).
- In 1966, the involvement of transglutaminase in cancer metastasis in a mouse model using the plasma cell tumour YPC-1 was proposed (Laki K, 1966).
- In 1989, the first microbial transglutaminase was produced by Streptoverticillium mobaraense.
- Transglutaminase was first developed and marketed industrially by Ajinomoto Co, Japan, by the name Activa TM TG and was utilised in food applications.
Since then the interest in this novel enzyme has been increasing. The major applications of transglutaminase will be discussed further in this literature review.
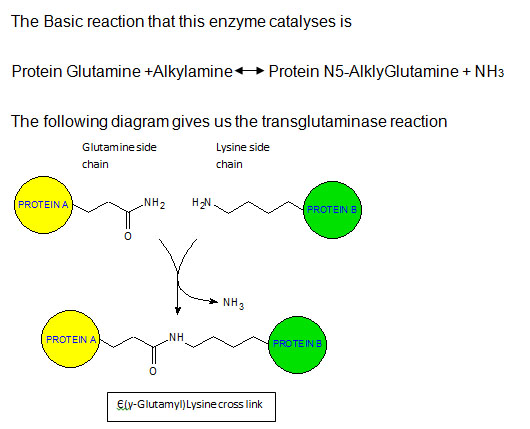
Transglutaminases are known as nature’s biological glues and industry’s biocatalysts. The enzyme’s function is to catalyse a strong covalent bond that is highly resistant to proteolytic degradation. The bond is formed between a free amine group (peptide bound lysine or protein) and the gamma carboxamid group of a protein or peptide bound glutamine. Transglutaminases are involved in the post- translation modification of proteins through acyl transfer reactions where peptidyl glutamine residues are the acyl donors and primary amines act as acyl acceptors. (Martin Griffin, 2002; Schleehauf, 2001). ‘Transglutaminases seem to react best with glutamine residues in unstructured flexible regions of proteins, often in the N and C terminal domains, but always in end positions’ (Kapil Mehta, 2005). In-vivo transglutaminase is said to be involved in cell proliferation, cell migration, wound healing apoptosis, ECM reorganization, receptor-mediated endocytosis.
Classification and types of transglutaminase
Transglutaminase (EC 2.3.2.13) is classified as follows
E.C. 2 Transferase
E.C. 2.3 Acyltransferase
E.C. 2.3.2 Aminoacyltransferase
E.C. 2.3.2.13 ProteinGlutamineGammaGlutamlytransferase
Transglutaminase is found in both eukaryotes and prokaryotes. Transglutaminase is present in many organisms including micro-organisms, invertebrates, plants, amphibians, fish and birds.
The most common sources of transglutaminase are mammals. Mammalian transglutaminase is calcium dependent whereas plant and bacterial transglutaminase is not calcium dependent.
Examples of mammalian transglutaminases are
| Transglutaminase | Function | Distribution | |
| Epidermal transglutaminase | Involved in cornified cell envelop formation during terminal differentiation of keratinocytes | 77kDa | Cytosol |
| Factor XIII | Associated with blood clotting and wound healing | 83kDa | Cytosol and extracellular matrix |
| Tissue transglutaminase | Involved in cell death, cell differentiation, matrix stabilisation and cell adhesion | 80kDa | Cytosol, nucleus, membrane, cell surface, extracellular |
| Keratinocyte transglutaminase | Involved in cornified cell envelop formation during terminal differentiation of keratinocytes | 90kDa | Cytosol, membrane |
| Prostrate transglutaminase | Involved in formation of rodent copulatory plug | 77kDa | Unknown |
(Martin Griffin, 2002)
As well as the above, three other types are present:
| TGM X | Found in skin |
| TGM Y | Unclear |
| TGM Z | Associated with testes and lungs |
Some other interesting facts about transglutaminase with respect to sequence alignment and structure:
Cysteine, histidine and aspartate (asparagines) are the three amino acids that comprise the catalytic triad. It has been found that the intermediate reaction is linked to the nucleophilic cysteine of the enzyme (Greenberg, C.S., 1994).It has also been found that papain-like thiol proteases share core structural folds with transglutaminase and exhibit similarities in two aspects; the reaction mechanism and in the catalytic triad. In 1999 (Kira S. Makarova, 1999), a study comparing and analysing a superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases was undertaken. It was found that the enzyme was comprised of three motifs: Motif I, containing the catalytic cysteine; Motif II, the active histidine; and Motif III, the active aspartate. The structure includes two strands for each of the three motifs and Motif I is associated with the α- helix. Two positions upstream of the catalytic cysteine another small residue is present and, also, unique aromatic residues are present in two positions downstream from the catalytic histidine and another aromatic residue at the N terminal side, flanking the catalytic aspartate. This small residue and the aromatic residues are conserved in animal and bacterial proteins. The two insert regions found inbetween these conserved motifs are different in archaeal, bacterial and animal transglutaminase.
Bacteria and archaea were found to have small inserts compared to animal species. On analysing the structure of Factor XIII using a multiple alignment, it was found that animal transglutaminase have two main inserts. The first insert present in animals was found to interact with the conserved β-strands and is positioned between Motif I and Motif II; it is a large structure compared to the inserts present in archaea and bacteria. The second insert is placed specially to interact with the C terminal domain of eukaryotic proteins (Kira S. Makarova, 1999).
There is an increasing interest in the inhibitors of transglutaminase. To name a few: cystamine, N-ethyl maleimide, iodoacetate, parachloromercuribenzoic acid, imidazoles and thiadiazoles. (G.A.H. DE JONG, 2003)
The problems associated with mammalian transglutaminase are:
- Limited supply
- Complicated downstream processing
- Calcium dependency and high molecular weight
- Expense
- Narrow substrate specificity
- Stable over a narrower pH and temperature range than microbial TGs
These are a few of the reasons why alternate sources of transglutaminase are being exploited. Research on microbial sources of transglutaminase is gaining popularity. Microbial transglutaminase, on the other hand, can be obtained easily by fermentation. It does not require calcium for activation and it could be produced in sufficient quantity.
Applications of transglutaminase
- Food industry
Transglutaminase is used to cross-link proteins. Usually it is used for meat binding and the joining of small chunks of meat into one big portion. In the meat industry transglutaminase is also used to improve low-grade meat which can be used to reduce overall cost. The following are some of the improvements that can be made in food properties:
- Obtaining a standardised texture.
- Improvement of breaking strength. This is particularly important for the mechanisation of meat preparation. It helps in the production of thin slices of meat.
- Water retention properties can be improved, making the meat juicier.
- Improved portion control.
- Substitution of other ingredients like salts and phosphates.
- Lower-grade portions can be used which cuts costs.
- Gelation capability of proteins can be improved.
- Viscosity of proteins and protein-containing products increases.
- Better thermal stability.
- Increased shelf life.
- Reduction in the allergenicity of food. This can be applied to wheat flour and gluten.
Microbial transglutaminase has a broad range of substrate reactivity. It acts on milk proteins (such as casein, sodium caseinate, β galactoglobulin, α lactalbumin), egg protein (egg white protein or obalbumin, egg yolk protein), meat protein (myoglobin ,collagen, gelatin, myosin and actin), soybean (11S globulin, 17S globulin), wheat (gliadin and glutenin)
Mostly mammalian transglutaminase has been used for all the above applications. The problem with mammalian transglutaminase is its low activity and high production costs. Therefore there is a need for a novel transglutaminase that can be produced industrially and can be utilised within the existing processes.
The putative transglutaminase proposed in this project can be used to obtain the above goal.
- Textile industry
Transglutaminase is used in the manufacture of woollen garments. Transglutaminase is added during wool processing while carrying out the scouring process or after carbonising. When transglutaminase is incorporated, the quality of the wool fibre improves drastically. Shrinkage and loss of strength is minimised with the addition of transglutaminase. Transglutaminase basically cross-links the scales present in the wool fibre. Antiparallel scales are made uniform by using transglutaminase that facilitates easy movement. There is an observable decrease in the total and felting shrinkage after treatment of wool with transglutaminase, making the garment shrink-proof and meeting consumer standards. Increase in the yarn strength is seen with the addition of transglutaminase. Enhancement in colour fastness after washing is seen. Dye loss is prevented and the original colour is retained when transglutaminase is applied to wool. Another enhancement with the application of transglutaminase to wool is that of the incorporation of functional compounds into wool fibres. These functional compounds can be amines including antimicrobial agents, anti-odour agents, dyes, etc. Fluorescein cadaverine can be easily incorporated into wool.
Thus transglutaminase prevents shrinkage, increase strength, reduces stretch, improves the way the garment feels and handles, and prevents dye loss.
- Leather industry
Bated bovine hide is a good substrate for transglutaminase. Mammalian transglutaminase has large molecules; they do not work very well in the leather industry as they face difficulties in being incorporated into the leather. For better incorporation of transglutaminase, there is the need for a microbial transglutaminase. This putative transglutaminase could provide an alternative source. Transglutaminase facilitates the ‘filling’ of leather and smoothes the irregularities present of the surface. Transglutaminase helps by cross-linking and thus providing more space for dye incorporation and opening up the leather for further processing.
Transglutaminase does not produce a tanning effect.
The leather structure is more open and transglutaminase increases resistance to collagenase and microbial attack.
- Medical applications
Transglutaminase (Factor XIII) is used in tisseal to form a ‘biological glue’. It is a mixture of fibrinogen, thrombin and Factor XIII. It is used in surgery to prevent blood loss and seal wounds. Transglutaminase is known to link fibrin molecules during the blood clotting process. Its use can reduce the need for stitches and contribute to the repair of fractures and cartilage lesions. In the liver, especially, this tisseal is a used to prevent blood loss. Transglutaminase is known to facilitate the scaffolding of proteins which is known to support further tissue healing and tissue re-formation (Yang Zhu 2008). In other words, this transglutaminase-facilitated scaffolding provides a durable matrix for cell attachment and further helps the spreading and proliferation of the cells involved. Thus transglutaminase has applications in tissue engineering.
The regulation of transglutaminase in mammals is very important. Recent studies have shown that transglutaminase plays a key role in certain chronic diseases. The cross-linking properties of this enzyme and its role in stabilisation reveals that any changes (down regulation/ negative control) in the regulation of this enzyme can lead to inflammatory diseases , ‘auto-immune conditions, chronic degenerative diseases like arthritis, atherosclerosis and neurodegenerative pathologies’ (Martin Griffin, 2002) like Huntington’s chorea, Alzheimer’s disease, and Parkinson’s disease. Transglutaminase is also involved in the treatment of cancer and tumours as it plays a role in cell death and apoptosis.
Geobacter sulfurreducens
Geobacter sulfurreducens is a metal-reducing anaerobic bacterium. It was isolated in a hydrocarbon-rich mine in Norman, Okla.Geobacter sulfurreducens measures about 2µm by 0.5 µm and is rod-like in shape. It is grown invitro on a defined medium containing acetate as the electron donor. Fumarate, malate, ferric citrate can act as electron acceptors. Optimum temperature for growth is around 35° C. The c-type cytochromes are observed in whole cells.
Its classification is as follows:
Super Kingdom: Bacteria
Phylum: Proteobacteria: Bacteria classified under this phylum are gram negative having outer membranes composed of lipopolysaccharides. Most of them have flagella required for their motility and pilli like structures for attachment. They are either chemoautotrophs, heterotrophic or facultatively or obligately anaerobic.
Subphylum: Delta/epsilon subdivions
Class: Deltaproteobacteria
Order: Desulfuromonadales
Family: Geobacteraceae
Genus: Geobacter
A previous study conducted by Janet Rollefson, Caleb Levar and Daniel Bond (Janet B. Rollesfson, 2009) has revealed that this bacterium is involved in microbial fuel cells. Previous research has shown that Geobacter sulfurreducens is capable of generating power in microbial fuel cells. Geobacter sulfurreducens is used mainly for:
- ‘Its genome sequence is available, making its study easier.
- It has a robust genetic system, making it easy to carry out the experimental procedures’ Applying Mini-Himar transposon mutagenesis, the researchers identified various mutants. GSU3361 mutant showed peculiar transglutaminase-like activity. This particular mutant showed a high attachment phenotype with respect to biofilm formation. ‘Growth and respiration was found to be identical to the wild type, followed by a sudden decrease and further death of the cells’ (Janet B. Rollesfson, 2009).
This bacterium is our research organism whose genome has been sequenced previously. We shall be looking at the putative transglutaminase gene to check whether it encodes for transglutaminase or not. If successful and rightly proposed this novel enzyme can be marketed and will find applications as described previously.
Transglutaminase is found to be associated with sporulation in bacteria and is involved in assembling the spore coat proteins. Transglutaminase is found to be localised on the spores of bacteria especially in the Bacillus species, namely Bacillus subtilis, Bacillus cereus, etc.(Kobayashi K, 1998). During the process of sporulation, there are two point of regulation; firstly, the production and expression of several proteins involved in coat formation and, secondly, the assembly of these coat proteins to form a rigid structure that is resistant to degradation by other proteolytic enzymes. Transglutaminase is thus found to play in role in cross-linking the spore coat proteins and forming rigid supra-molecular structures (Katerina Ragkousi 2004).
Geobacter species are found to play a role in environmental restoration. They have been used to feed on oil spills and ground-water pollutants to clean the environment.
References
CLARKE D. D, M. M. J., NEIDLE A. AND WAELSCH H. 1957. The incorporation of amines into proteins Arch Biochem Biophys, 79, 338-354.
G.A.H. DE JONG, G. W., AND S.J. KOPPELMAN 2003. Transglutaminase Inhibitor from Milk. JOURNAL OF FOOD SCIENCE, 68, 820-825.
GREENBERG C.S., H. J. M. 1994. Analysis of the catalytic activity of human factor XIIIa by site-directed mutagenesis journal of biological chemistry, 269, 28309-28313.
JANET B. ROLLEFSON, C. E. L., AND DANIEL R. BOND 2009. Identification of genes involved in biofilm formation and respiration via Mini-Himar transposon mutagenesis of Geobacter sulfurreducens. Journal of Bacteriology, 191, 4209-4217.
KAPIL MEHTA, R. E. 2005. Transglutaminases, Family of enzymes with diverse function. Progress in Experimental Tumor Research, 38, 1-265.
KATERINA RAGKOUSI , P. S. 2004. Transglutaminase-Mediated Cross-Linking of GerQ in the Coats of Bacillus subtilis Spores Protein Science, 8, 1714-1719.
KIRA S. MAKAROVA, L. A. A. E. V. K. 1999. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminase. Protein Science, 8, 1714-1719
KOBAYASHI K, S. S., IZAWA Y, MIWA K, YAMANAKA S 1998. Transglutaminase in sporulating cells of Bacillus subtilis. The journal of general and applied microbiology, 44, 85-91.
LAKI K, T. H., YANCEY ST 1966. Clot forming and clot stabilizing enzymes from the mouse tumor YPC1. Biochem Biophys Res Commun, 22, 776-781.
MARTIN GRIFFIN, R. C. A. C. M. B. 2002. Transglutaminase: Nature’s Biological Glue. Biochemical Journal, 368, 337-396.
PISANO, J. J., FINLAYSON, J. S AND PEYTON, M. P 1968. Cross-link in fibrin polymerizes by Factor 13: e-(y-glutamyl)lysine. Science, 160, 892-893.
SCHLEEHAUF, S. 2001. A novel microbial transglutaminase derived from Streptoverticillium baldaccii. Doctor of philosophy, Griffith University.
YANG ZHU , J. T. 2008. Novel applications for microbial transglutaminase beyond food processing. Trends in Biotechnology, 26, 559-565.